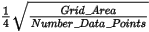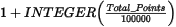|
Low-pass
desampling factor
|
The de-sampling factor is defined as a function of the grid cell size. Before any further calculations, all points within cells of dimension cell_size x desampling_factor are averaged into a single value and placed in the center of the cell.
The default desampling (desampling_factor) is set relative to the contributing points as:
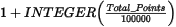
Desampling has two impacts:
If the database contains less than 100000 contributing points, the default is set to 1, and the only pre-filtering consists of de-aliasing at the cell level.
The variogram calculation duration is a function of the square of the contributing points. Increasing this factor visibly cuts down the variogram calculation time while little impact is seen on the variogram curve.
Script Parameter: E3MGRGRD.DSF
|